Identification of the source of donor derived cell-free DNA in a kidney-pancreas transplant recipient: case study
Renata Ponsirenas1, Natali Gulbahce2, Evgeniya Vaskova3, Kim Langone3, Marica Grskovic3, George W Burke III4.
1Medical Affairs, CareDx, South San Francisco, CA, United States; 2Data Sciences, CareDx, South San Francisco, CA, United States; 3R&D, CareDx, Brisbane, CA, United States; 4Department of Surgery, University of Miami, Jackson Health System, Miami, FL, United States
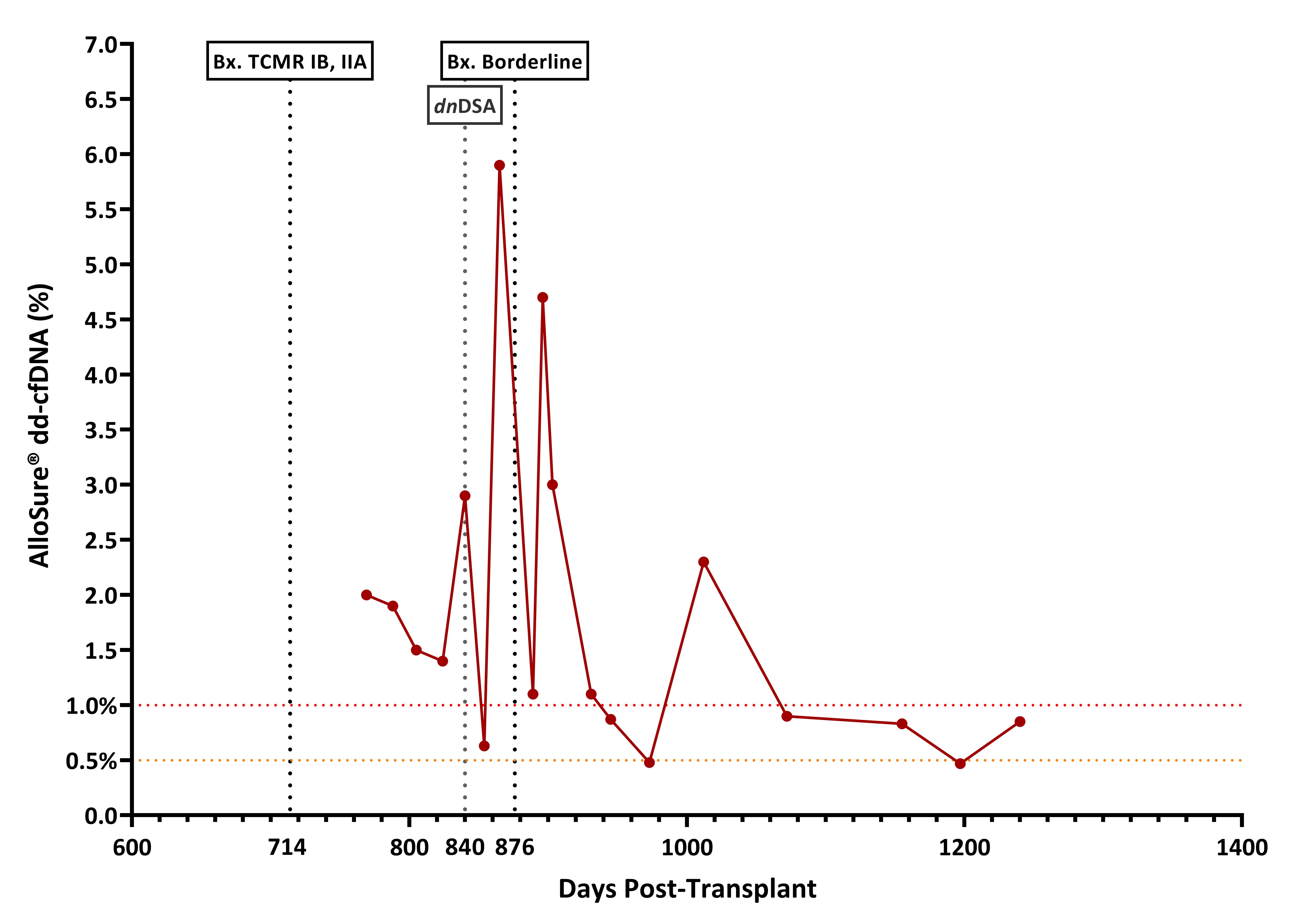
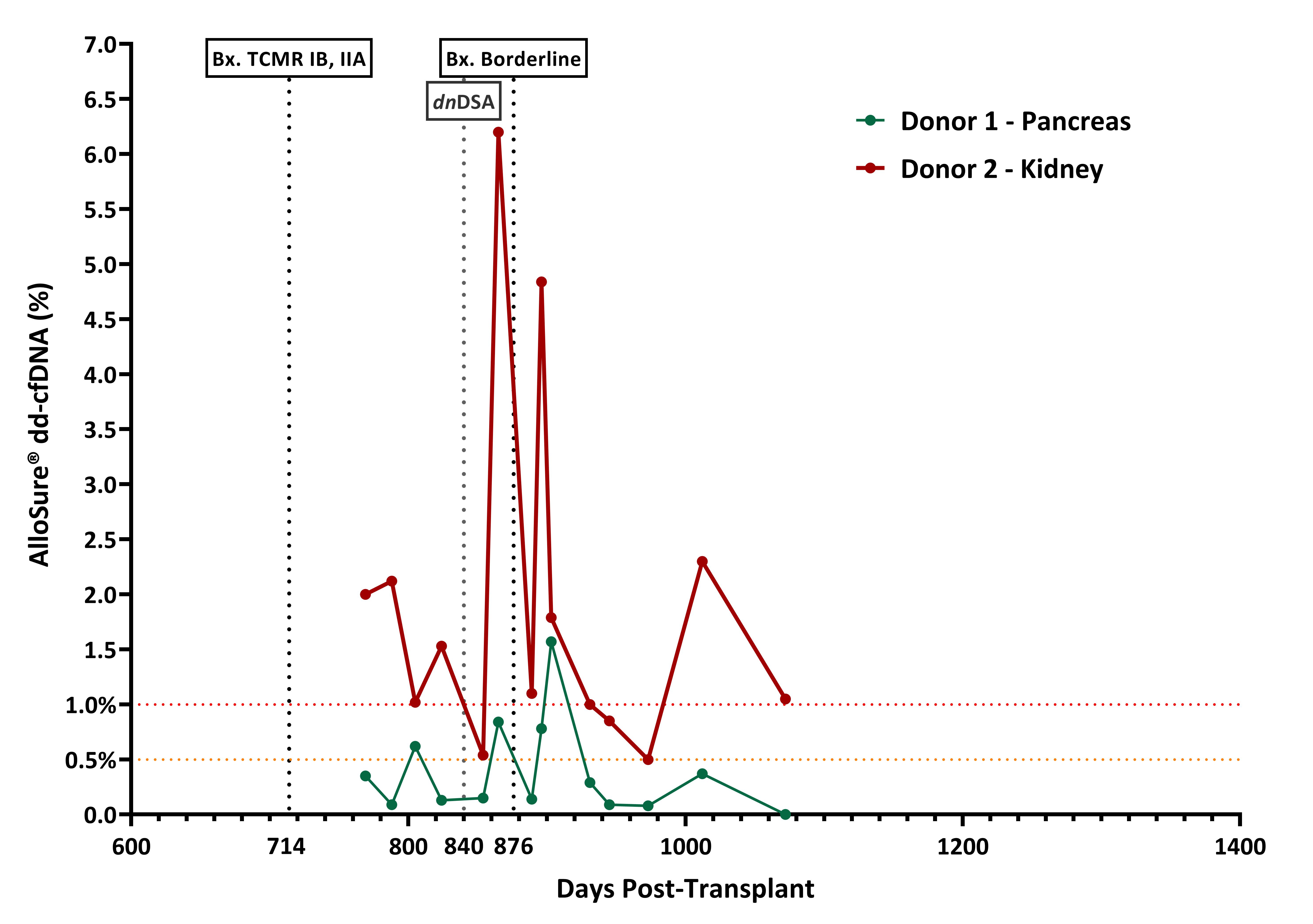
Although outcomes of Kidney-Pancreas Transplant (KPT) have steadily improved over the years rejection remains the primary cause of graft loss after 3 months. The surveillance for graft rejection is done by serum creatinine, for the kidney, and pancreatic enzymes for the pancreas. Both can be non-specific to graft injury. Donor derived cell-free DNA (dd-cfDNA) is a non-invasive biomarker recently added to the standard of care of many transplant centers to monitor for rejection in Kidney Transplant (KT) recipients. The use of dd-cfDNA in KPT brings some challenges since it is typically not possible to identify organ source of elevated dd-cfDNA without genotype information. Additionally, when a second KT is necessary after the failure of a first kidney, in the context of keeping the pancreas, then genotypes of both donors and recipient are typically required to identify organ source. Here we report the application of a novel algorithm that does not require genotyping of both donors, to evaluate the levels of each dd-cfDNA in a KPT recipient who had a second KT. A 50-year-old female received a KPT in 2012 from a deceased donor. Since this was a bladder drained pancreas transplant, urine amylase was followed in addition to creatinine and pancreatic enzymes. Patient developed kidney chronic rejection and received a second living donor KT in 2018 with removal of the original KT in the same operation. The pancreas transplant continued to function well. 2 years later a biopsy of the second KT demonstrated TCMR IB, IIA. After treatment with steroids pulse, patient was surveilled with AlloSure® dd-cfDNA. Although, no clinical signs of rejection, dd-cfDNA showed high variability over time (Figure 1). A second kidney biopsy was performed showing borderline rejection. Patient was treated with steroids pulse but developed donor specific antibodies (DSA). To avoid a pancreas biopsy, we obtained DNA material from the recipient and from the donor of the second KT to determine if the elevated levels of dd-cfDNA was from injury to the pancreas. A novel algorithm was used to differentiate the SNPs (Single Nucleotide Polymorphisms) from donor 1 (1st KPT and donor 2 (KT). We were able to determine that most dd-cfDNA was being released by the 2nd kidney transplant, but both organs showed high variability over time (Figure 2). High variability in dd-cfDNA serial results is usually linked with non-adherence or improper intake of immunosuppressive drugs. Also, the presence of de novo DSA is associated with increase in dd-cfDNA and both combined over time with poor long outcomes. There is a need for more specific non-invasive biomarkers in dual organ transplantation. dd-cfDNA in surveillance of rejection in KPT has shown to be a useful tool. The ability to identify the specific organ source of the injury allows for the reduction of unneeded biopsies and better clinical management.