Development of high-performing and multicenter-validated dynamic prediction models with longitudinal measurements of serum creatinine and proteinuria for death-censored kidney graft failure
Soufian Meziyerh1, Anirudh Tomer2, Hessel Peters-Sengers3, Danny van der Helm1, Maarten Coemans4, Tobias Pieters5, Jan-Hendrik Venhuizen6, Saskia Haitjema6, Dirk Jan Moes7, Wieneke Michels1, Johan de Fijter1, Paul van der Boog1, Tri Nguyen8, Sandrine Florquin9,10, Arjan van Zuilen5, Frederike Bemelman11, Azam Nurmohamed, Ewout Steyerberg, Dimitris Rizopoulos, Maarten Naesens, Aiko de Vries, Jesper Kers.
1Leiden Transplant Center, Leiden University Medical Center, Leiden, Netherlands; 2Dept. of Biostatistics, Erasmus University Medical Center, Rotterdam, Netherlands; 3Center for Experimental and Molecular Medicine, Amsterdam University Medical Centers, Amsterdam, Netherlands; 4Dept. of Nephrology and Renal Transplantation, University Hospitals Leuven, Leuven, Belgium; 5Dept. of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, Netherlands; 6Utrecht Patient Oriented Database, University Medical Center Utrecht, Utrecht, Netherlands; 7Dept. of Clinical Pharmacy and Toxicology, Leiden University Medical Center, Leiden, Netherlands; 8Dept. of Pathology, University Medical Center Utrecht, Utrecht, Netherlands; 9Dept. of Pathology, Amsterdam University Medical Centers, Amsterdam, Netherlands; 10Amsterdam Infection & Immunity Institute, University of Amsterdam, Amsterdam, Netherlands; 11Renal Transplant Unit, Amsterdam University Medical Centers, Amsterdam, Netherlands; 12Dept. of Public Health, Erasmus University Medical Center, Rotterdam, Netherlands; 13Dept. of Biomedical Data Sciences, Leiden University Medical Center, Leiden, Netherlands; 14Dept. of Microbiology, Immunology and Transplantation, KU Leuven, Leuven, Belgium; 15Van 't Hoff Institute for Molecular Sciences, University of Amsterdam, Amsterdam, Netherlands
Introduction: Prediction models of graft survival in kidney transplantation allow risk stratification of kidney transplant recipients using big data and artificial intelligence. Many models require detailed information on allograft histology and presence of donor-specific antibodies which are not readily available in every transplant center. We developed a joint model using Bayesian estimation to correlate baseline transplant characteristics and longitudinal measurements of both serum creatinine (S-Cr) and urine protein-creatinine ratios (u-PCR) with death-censored graft failure (DCGF).
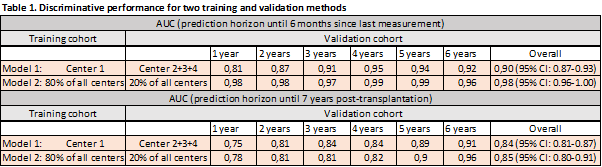
Methods: 3596 kidney transplant recipients (KTRs) from four transplant centers in Europe were included with a total of 452.033 S-Cr and u-PCR measurements. Each KTR had a median of 112 (IQR: 52-163) consecutive measurements. The interval between measurements was 7 (IQR: 1-35) days. Median follow-up was 6 (IQR: 3-9) years. Model predictors included recipient and donor gender and age, total number of human leukocyte antigen-mismatches, donor type, pre-transplant DSAs, and dialysis vintage. Primary outcomes were DCGF six months after last S-Cr or u-PCR measurement and DCGF seven years post-transplantation. Two training and external validation methods have been used to assess best model performance: model 1; development in one center and validation in three remaining centers, and model 2; development on 80% of the total cohort and validation on the remaining 20% (Table 1).
Results: A total of 549 (15%) out of 3596 KTRs experienced DCGF. Joint modeling revealed that recipient age, recipient gender, donor age, transplantation after dialysis initiation, and serial measurements of S-Cr and u-PCR were independent risk factors for DCGF. Our final joint models showed good calibration (prediction error: 0.01) and very high discrimination in the development and validation cohorts (6-months incident AUC 0.9 [95% CI: 0.87-0.93] and 7-years dynamic AUC 0.84 [95% CI: 0.81-0.87] for model 1, and 6-months incident AUC 0.98 [95% CI: 0.96-1.00] and 7-years dynamic AUC 0.85 [95% CI: 0.80-0.91] for model 2) (Table 1). Especially model 2 could be of interest as a personalized surveillance tool in clinical practice given its AUCs >0.95 from year 1 onwards.
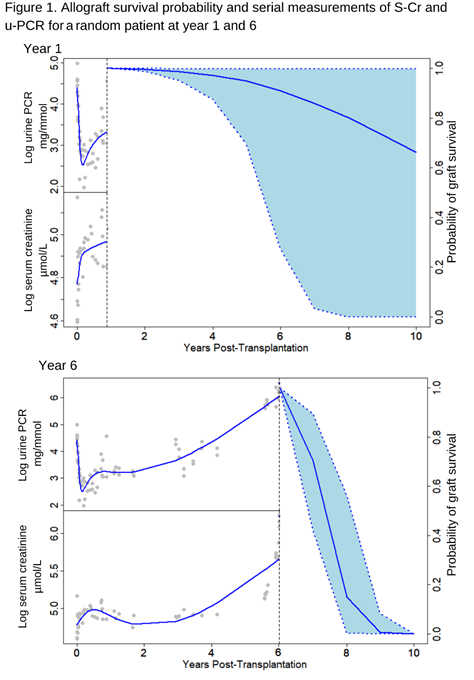
Conclusion: We developed a high performing and multicenter-validated, dynamic prediction model on death-censored allograft survival based on traditionally available baseline transplant characteristics and longitudinal measurements of S-Cr and u-PCR. This dynamic model can continuously be updated with new measurements and might enable personalized surveillance of kidney transplant recipients and objectify allograft prognosis (as illustrated in Figure 1). Of notice, our models can be extended with newly discovered dynamic biomarkers in the future if needed.