Unravelling the molecular footprint of kidney graft quality: from subjective assessment to tailored treatment
Leonie van Leeuwen1,2, Darragh P. O'Brien2, Asel Arykbaeva3, Ian Alwayn3, Dorottya de Vries3, Cyril Moers1, Rutger Ploeg1,3,4, Henri G.D. Leuvenink1, Benedikt M. Kessler2.
1Department of Surgery, University Medical Center Groningen, Groningen, Netherlands; 2Nuffield Department of Medicine, Centre for Medicines Discovery, Target Discovery Institute, University of Oxford, Oxford, United Kingdom; 3Department of Surgery, Leiden University Medical Center, Leiden, Netherlands; 4Nuffield Department of Surgical Sciences, University of Oxford, Oxford, United Kingdom
Individual kidney grafts are exposed to damage in different ways (e.g., donor type, donor age) and it is likely that the underlying molecular mechanisms differ. In addition, 20% of donor kidneys are judged irreparably damaged and discarded, primarily due to lack of knowledge regarding the underlying pathophysiological mechanisms. Knowledge of protein degradation leading to loss of tissue integrity and function is particularly scarce. We therefore sought to gain molecular insights into the role of protein (degradation) dependent mechanisms associated with different donor characteristics of kidney grafts.
Renal cortex biopsies were collected from deceased donor kidneys (n=15), which were declined for transplantation due to medical and logistical reasons. Biopsies, collected after cold preservation, were analyzed using a combined proteomics and degradomics approach in a non-biased manner. Kidney grafts were classified based on donor characteristics (Table 1). PANTHER pathway enrichment and STRING functional analysis were used to link proteins to biological processes. TopFind analysis was used to match degradation products to their associated protease.
" href="https://cm.tts2022.org/papers/body/1008#">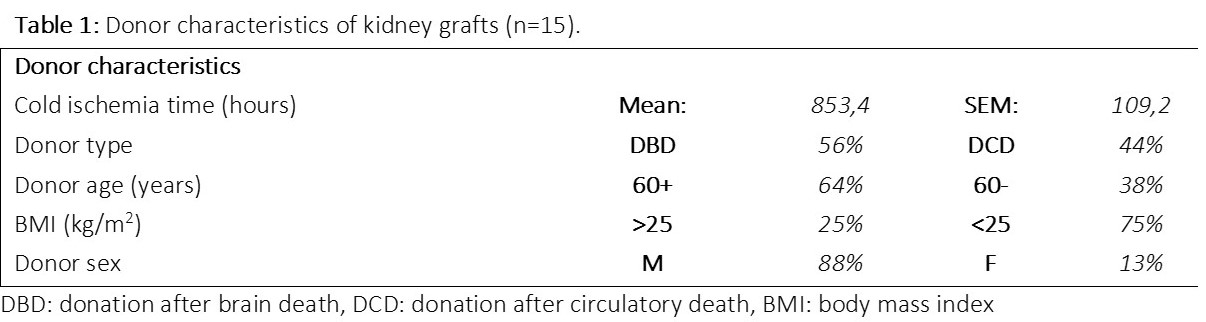
Kidneys from brain dead donors showed enrichment of respiration and energy related processes, and degradation occurred mostly in plasma related proteins residing in tissue material (Figure 1). Kidneys from donors after circulatory death revealed degradation of structural proteins. Older donors exhibited protein enrichment of processes related to immune response and complement activation, but degradation of apoptotic related proteins and proteins responsible for cytoskeletal cleavage. Conversely, younger donors revealed enrichment of mitochondrial and aerobic electron transport related processes, while degradation mostly occurred in proteins related to cell binding. Donors with a BMI of <25 also exhibited enrichment of respiration and energy related processes, but degradation of antioxidants and proteins related to cytoskeletal organization. Donors with an BMI of >25 showed degradation of proteins related to fatty acid metabolism, degradation processes and ATP synthesis. Male donors revealed enrichment of processes related to immune response and complement activation, however showed degradation of mitochondrial proteins. Female donors showed degradation of proteins related to the proteasome complex, and structural proteins.
Pathway enrichment analysis revealed different biological pathways upregulated depending on donor characteristics, confirming unique molecular footprints of kidney grafts. Furthermore, proteome and degradome pathways were not always aligned, indicating a potential complex interplay between protein dysregulation and proteolysis. These results reveal novel insights into the pathophysiological mechanisms of deceased donor kidney quality and could lead to novel drug targets for tailored treatment of donor kidney grafts.
" href="https://cm.tts2022.org/papers/body/1008#">
Proteomics experiments were funded by Novo Nordisk, awarded to Benedikt Kessler. Machine perfusion experiments were funded by Dutch Kidney Foundation awarded to Ian Alwayn as part of the clinial PROPER trial (NCT04693325). Travel to Oxford was supported by Stichting de Drie Lichten and the Graduate School of Medical Sciences of the University of Groningen, awarded to Leonie van Leeuwen.

right-click to download
