Combined genome wide and single nuclei transcriptomics in posttransplant acute kidney injury identifies early proinflammatory/profibrogenic cell origin
Jennifer McDaniels1, Amol Shetty2, Elissa Bardhi1, Thomas Rousselle1, Enver Akalin3, Daniel Maluf4, Valeria Mas1.
1Department of Surgical Sciences, University of Maryland School of Medicine, Baltimore, MD, United States; 2Institute of Genomic Sciences, University of Maryland School of Medicine, Baltimore, MD, United States; 3Albert Einstein College of Medicine, Montefiore Medical Center, Bronx, NY, United States; 4Department of Surgery, University of Maryland School of Medicine, Baltimore, MD, United States
Introduction: Kidney transplants offer a unique opportunity to evaluate molecular pathways involved in development and response to acute kidney injury (AKI) as most patients experience some level of AKI and graft biopsies are done frequently. This prospective study of transplanted kidneys with AKI investigates their unique transcriptomic profile and their role in chronic allograft dysfunction.
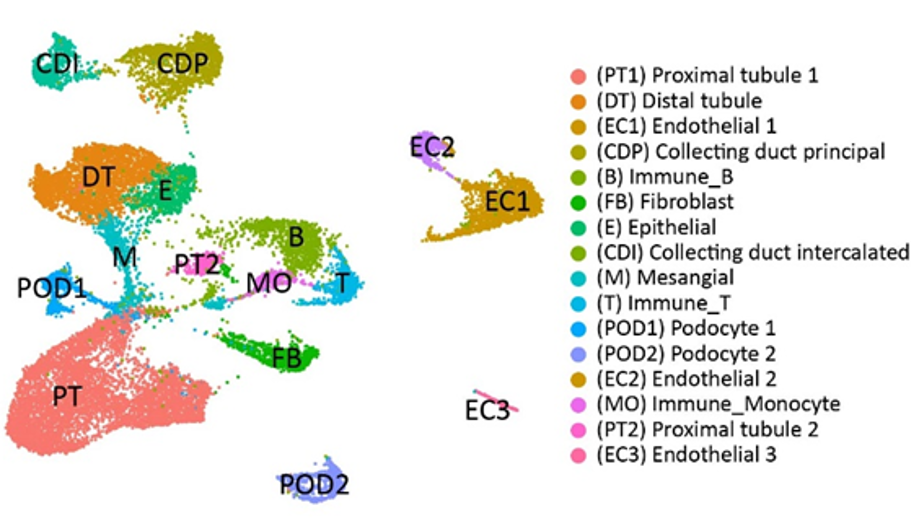
Methods: Fifteen AKI biopsies with histological features of acute tubular injury (pure-AKI) (collected within 1-month posttransplant) and paired 3-month protocol biopsies (n=30 biopsies) were assayed by microarrays. Normal allografts (NAs) biopsied at 3-months posttransplant were used as controls (n=20). AKI patients were subclassified based on their graft function (high/low) at 36-months posttransplant. A small subset of AKI samples (n=4) and NA (n=3) were evaluated using single nuclei(sn) RNA-seq to identify cells involved in injury or impaired reparation. Using the 10X Genomics Chromium Platform, >40,000 nuclei in Gel-Bead V3 were captured. Analysis of each dataset was generated with the CellRanger software and visualized using UMAP. Distinct cell clusters were identified by their expression of highly variable genes.
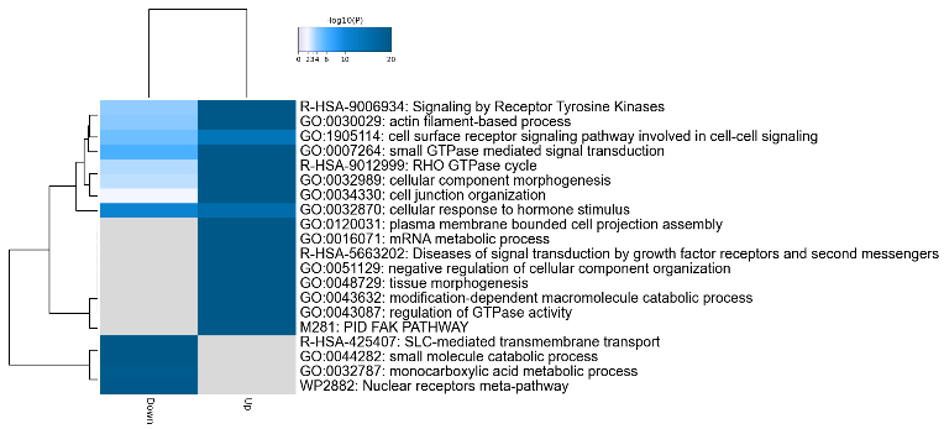
Results: DEGs (FDR<0.05) between pure-AKI and paired 3-month biopsies showed upregulation of the humoral immune response, neutrophil degranulation, and wound healing in kidney grafts with eventual low function at 36-months posttransplant. Gene enrichment analyses showed upregulation of epithelial mesenchymal transition, angiogenesis, and collagen formation in the same biopsies. Following snRNA-seq, 16 cell clusters were identified from pure-AKI and NA samples (UMAP displayed in Fig 1). Two types of proximal tubule cells were identified (PT1 and PT2) and characterized by decreased metabolic functions and SLC-mediated transmembrane transport in AKI grafts. The top up- and down-regulated PT biological pathways in AKI are shown in a heatmap in Fig 2. PT2 was characterized by dedifferentiation of PT. Three subsets of endothelial cells (EC1-3) were also identified. EC1 presented markers of glomerular capillary ECs. Fibroblasts presented an activated phenotype compared to NAs (extracellular matrix turnover, collagen biosynthesis). Proinflammatory monocytes were predominant in AKI samples.
Conclusion: DEGs between paired pure-AKI and 3-month biopsies associated with immune activation and impaired reparation in kidney grafts in allografts with low function at 36-months posttransplant. Critically, the analyses of transcriptional profile at single cell resolution identified early proinflammatory/profibrogenic cell subtypes. Further evaluation of cell subtypes and cell-to-cell interactions may lead to the discovery of targeted interventions to avoid progression to fibrosis and graft function loss.

right-click to download
