Genetic identity evaluation in human cell cultures for clinical research/cell therapies: comparison of two molecular methods
Gustavo Ariel Wildfeuer1, Daniela Patricia Fernandez Souto1, Cecilia Gamba1, Silvina Kuperman1, Valeria Roca1.
1Centro Regional de Hemoterapia, Hospital de Pediatría SAMIC “Prof. Dr. J. P. Garrahan”, CABA, Argentina
Introduction: Cell therapies are a constantly growing field of research, with benefits documented for numerous conditions in clinical and pre-clinical studies. Within these studies, cell expansion or cell culture is often needed. Cell line contamination and misidentification is a significant threat facing cell culture, with the potential to invalidate resulting data.
To avoid misidentification in Human cell lines for research, performing short tandem repeat (STR) profiling is recommended. However, for cell therapies products guidance only states that testing procedures should be implemented to ensure cell cultures identity and that acceptable limits for culture composition should be defined.
The National Public Umbilical Cord Blood Bank of the Garrahan Pediatric Hospital (BSCU) executes quality control of the manufacturing process as well as the final product according to national and international standards and regulations. HLA typing, by means of PCR Sequence-Specific Oligonucleotide (SSO) technique (LABType One Lambda), is commonly used in many laboratories to asses compatibility for trasplantation, including ours. HLA genes are highly polymorphic, hence we aimed to study whether HLA typing by SSO-PCR could be used as a method to asses genetic identity in cell cultures intended for cell therapies.
Method: DNA samples (5) were obtained from HSC of 4 umbilical cord blood and 1 peripheral blood from the mother of one of them after Informed Consents were signed. Mixed DNA serial dilutions (1:10; 1:25; 1:50) were analyzed for genetic identity by both molecular methods. STR (AmpFLSTR identifiler Plus, Applied Biosystems) was performed according to manufacturer's protocols and PCR-SSO technique (LABType, One Lambda) was performed according to SOP.
Results: Unique DNA profiles were obtained for each sample with both methods. For the DNA mixtures, we were able to detect contamination in all the conditions (100%) analyzed by STR profiling. However, HLA typing showed different results depending on the gene analyzed (Table 1).
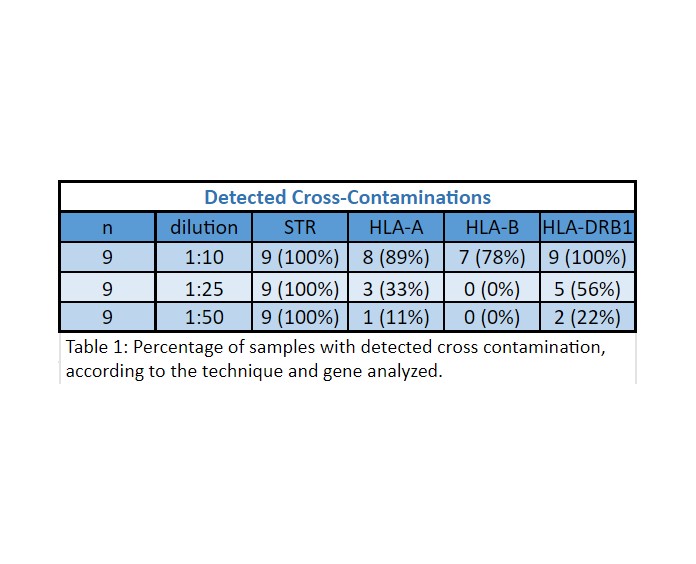
Conclusion: HLA typing was successful to detect cross contaminations with higher sensitivity at HLA-DRB1 gene (1:10). At the other loci under study, sensitivity varied depending on the analyzed gene and contamination degree.
STR was able to detect all of the contaminated samples, despite the contamination levels.
Since the scientific/clinic community is yet to determine which is the degree of cross-contamination of a culture that could negatively influence cell therapy or clinical research, both techniques could be used depending on each laboratory capabilities.
