Single cell RNA sequencing of cryopreserved human renal transplant core biopsies: a feasibility and optimisation study
Oliver McCallion1, Matthew Brook1, Joanna Hester1, Fadi Issa1.
1Nuffield Department of Surgical Sciences, University of Oxford, Oxford, United Kingdom
Background: Single cell RNA sequencing (scRNAseq) of tissue infiltrating leukocytes within human kidney transplants presents several technical challenges. For example, dissociation protocols may cause cell death or transcriptomic skewing, whilst creating single cell suspensions from freshly acquired biopsies is often impractical.
Aims: 1) compare and optimise leukocyte yield from three dissociation protocols and 2) evaluate the feasibility of scRNAseq on leukocytes from cryopreserved human kidney transplant biopsies.
Methods: Three digestion protocols (cold, warm manual, and warm automated enzymatic digestion) were evaluated using core biopsies from porcine kidney to optimise dissociation protocols. Following this, renal biopsies were taken from two patients at 5- and 9-months following transplant and dissociated into single cell suspensions by warm manual enzymatic digestion, which were then flow sorted to isolate CD45+ cells. cDNA libraries were produced with either 3’ or 5’ gene expression (GEX) kits from 10x Genomics and sequenced by Illumina NextSeq. Reads were aligned to the reference transcriptome GRCh38 (CellRanger) with QC, normalisation (SCTransform), clustering, and annotation performed with Seurat and SingleR.
Results: Warm manual enzymatic digestion was found to be the superior method for dissociation. With warm manual enzymatic digestion, a mean of 3906 (2512—6045) live CD45+ cells were isolated. Libraries were successfully generated for two biopsies: the 5-month biopsy from the first patient using the 3’ GEX preparation protocol; and the 9-month biopsy from the second patient using the 5’ GEX protocol. Following QC, a total of 12475 RNA transcripts in 396 cells, and 13128 RNA transcripts in 699 cells were identified in the 5-month and 9-month biopsy respectively. Clustering and automated annotation demonstrated that T cells, followed by NK cells, were the most abundant cell types identified.

Of the top 20 highly variable genes, 6 were common to both samples (GNLY, HLA-DRA, CCL4, GZMB, CST3, and AIF1). Allograft Inflammatory Factor 1 (AIF1) has previously been identified as a marker of chronic rejection and vascular inflammation whilst Cystatin 3 (CST3) is a biomarker of kidney function. Expression of both these genes localised specifically to the monocyte/macrophage cluster within both samples.
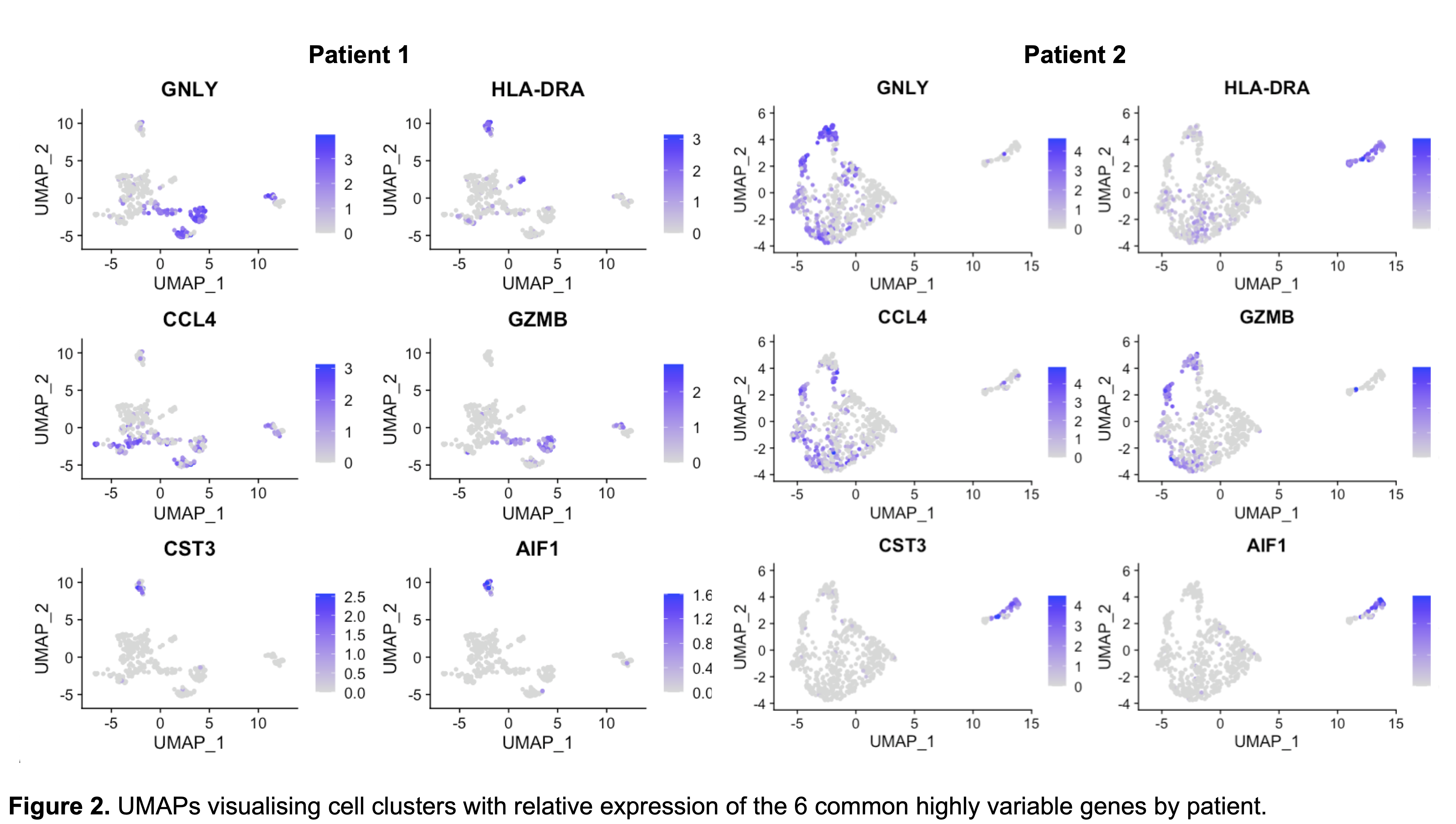
Discussion: Here we successfully characterise the leukocyte transcriptome within cryopreserved human kidney core biopsies. We identify an inflammatory monocyte/macrophage signature within samples dissociated by this protocol. Whether this is biological or technical remains to be determined, particularly as both patients maintain stable transplant function. Nevertheless, we demonstrate that scRNAseq from cryopreserved human transplant biopsies is feasible and enables deep transcriptomic analysis of tissue-infiltrating immune populations.
OM is an Medical Research Council Clinical Research Training Fellow (MR/V000942/1). FI is a Wellcome Trust CRCD Fellow (211122/Z/18/Z). The work on advanced cellular therapies in the authors group is supported by the UK Medical Research Council (MR/N027930/1) and the European Union’s Horizon 2020 research and innovation program under grant agreement No 825392 (ReSHAPE).
