The molecular nature of the banff iIFTA lesion
Harry Robertson1,2, Sebastian Hultin1, Jennifer Li1,3, Brian Nankivell3, Natasha M Rogers1,3, Ellis Patrick1,2, Philip O'Connell1.
1Centre for Transplant and Renal Research, Westmead Institute for Medical Research, Westmead, Australia; 2School of Mathematics and Statistics, University of Sydney, Sydney, Australia; 3Department of Renal Medicine, Westmead Hospital, Westmead, Australia
Introduction: Inflammation within areas of interstitial fibrosis and tubular atrophy (iIFTA) have been linked to adverse outcomes in kidney transplantation through an association with T-cell immunity leading to altered renal parenchymal structures. To date there is no literature that describes changes in the transcriptome in transplant recipients with iIFTA lesions. Further, molecular evidence for the progression of the banff iIFTA lesion, within the first year of transplantation, remains absent from the literature. Herein we define molecular changes within kidney allografts with an iIFTA lesion. We further characterise molecular signatures that accurately describe the progression of iIFTA at 12-months on a 3-month protocol biopsy.
Methods: We performed RNA sequencing of 113 protocol biopsies with all samples scored by the same histopathologist. Differentially expressed genes were deemed significant if they had a Benjamini-Hochberg adjusted P < 0.05. We then performed a gene set enrichment analysis using the gene ontology database to identify biological pathways that were enriched in iIFTA lesions. We also compared the transcriptomic profiles of patients with antibody- and T-cell mediated rejection with the iIFTA lesion. We further interrogated the same cohort for the progression of their iIFTA score at 12-months. Differential gene expression, and pathway analysis were employed to identify risk genes in patients that developed high iIFTA at 12-months post-transplant.
Results: Of the 113 biopsies with RNA sequencing, 37 demonstrated iIFTA by histopathology. Following transcriptomic analysis, 336 genes were differentially expressed in biopsies with an iIFTA diagnosis. The top tanking genes and pathways were all involved in the formation of immunoglobin and B-cell activation. Further, transcriptomic changes in the iIFTA lesion closely resembled those found in antibody-mediated rejection. Finally, we demonstrate that genomic information, from both peripheral blood and kidney biopsy 3-months post-transplant, provides better prediction power than clinical data in identifying patients with iIFTA lesions. 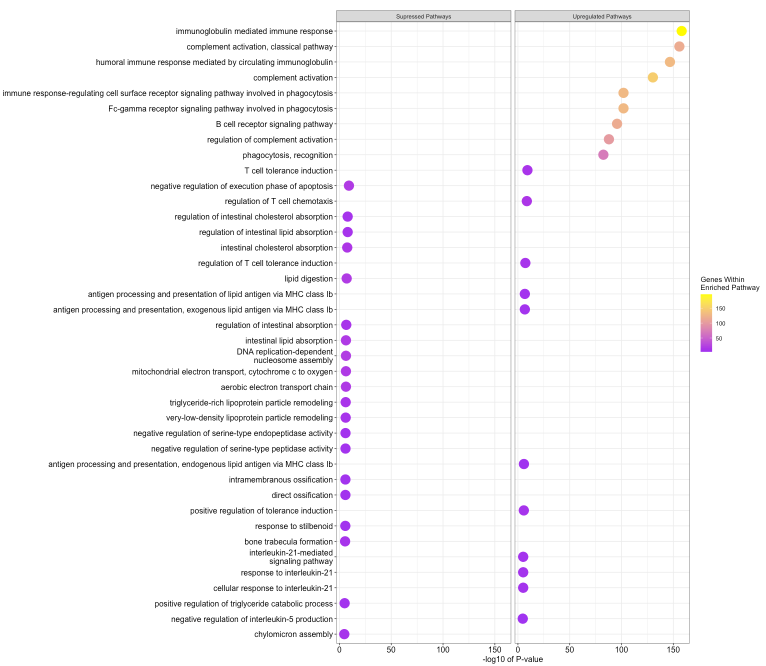
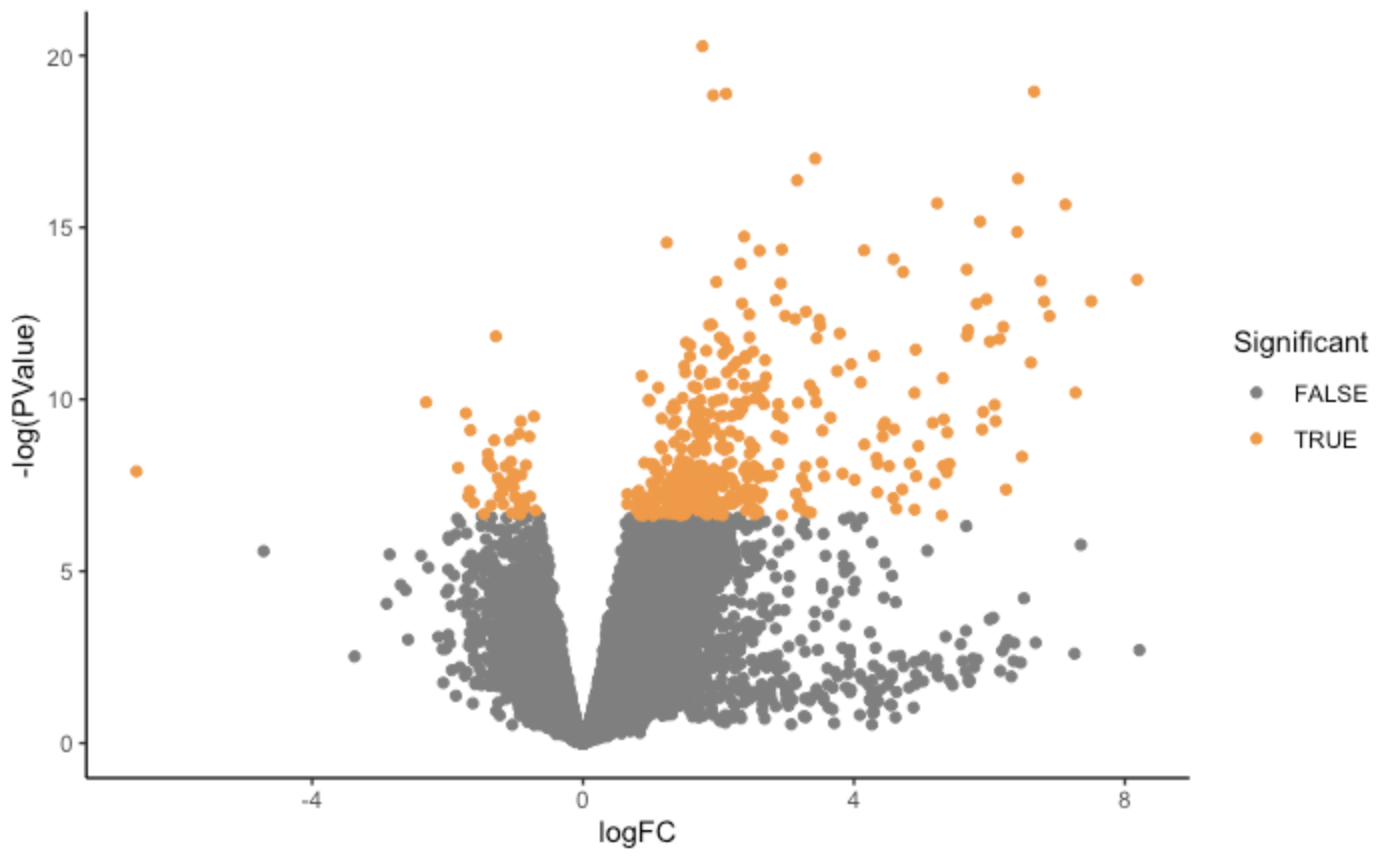
Conclusion: RNA sequencing enabled us to identify novel transcriptomic changes in patients with iIFTA. This pathology is primarily driven by immune-based changes, closely resembling antibody-mediated rejection, which is at odds with the current concept of iIFTA mediated by T-cell activation. Further, this study aims to build a foundation for using molecular technology in identifying the progression of the Banff iIFTA lesion.
