Moumita Paul-Heng, Australia has been granted the TTS-TSANZ International Transplantation Science Mentee-Mentor Awards
Single cell alloreactive TCR repertoire profiling
Moumita Paul-Heng1, Martina Denkova1, Eric Taeyoung Son1, Thomas Ashhurst2, Mario Leong1, Claerwen Jones3, Pirooz Zareie3, Anthony Purcell3, Nicole L La Gruta3, Nicole Mifsud3, Alexandra Sharland1.
1Transplantation Immunobiology Research Group, The University of Sydney, Sydney, Australia; 2Sydney Cytometry Core Research Facility, The University of Sydney and Centenary Institute, Sydney, Australia; 3Monash Biomedicine Discovery Institute, Monash University, Melbourne, Australia
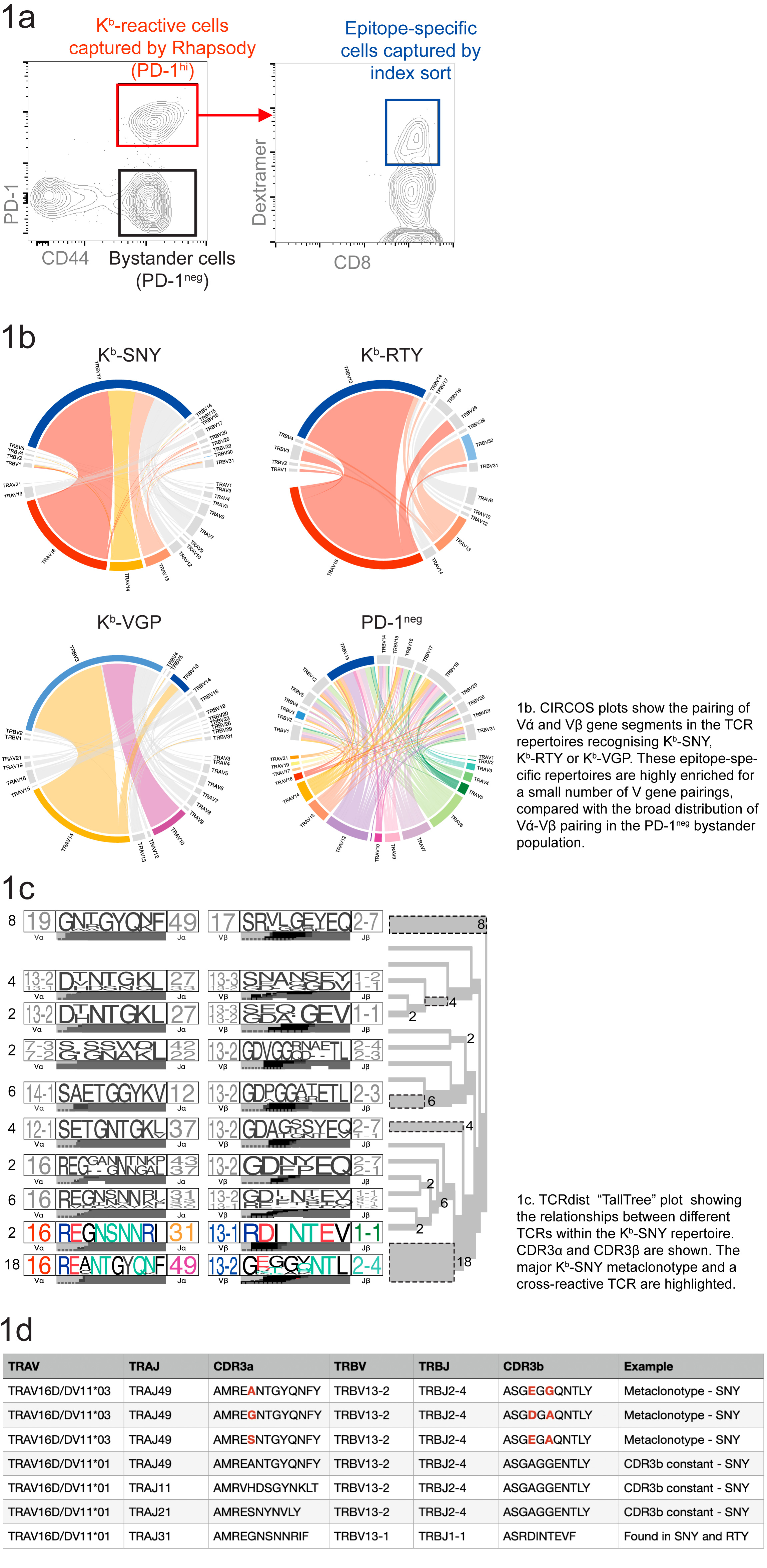
Aims: We discovered >40 Kb-peptide epitopes directly recognised by alloreactive CD8 T cells from B10.BR mice (H-2k). Here, we integrated two approaches to profile the alloreactive T cell repertoire at an early stage in tolerance induction (Fig.1a).
Methods: B10.BR mice were primed with a Kb-expressing skin graft followed by inoculation with AAV-Kb. Nested PCR and Sanger sequencing of paired αβ TCR from single dextramer-positive cells was performed in parallel with BD Rhapsody library preparation/Illumina sequencing for paired TCR and targeted transcriptome analysis.
Results: Repertoires for 3 dominant epitopes (Kb-SNY, Kb-RTY and Kb-VGP) were determined (Fig.1b). TCR αβ diversity was significantly reduced among epitope-specific T cells (scores Kb-SNY 47.3, Kb-RTY 69.5, Kb-VGP 279.5) compared with PD-1neg bystander cells (score 78730). Alloreactive repertoires were strongly skewed towards usage of particular V-J gene segments and comprised families of related TCRs including both private TCR clones and public meta-clonotypes (Fig.1b-d). One cross-reactive clone recognised both Kb-SNY and Kb-RTY (Fig.1c-d). A number of clones shared the same b chain paired with different alpha chains (Fig. 1d) while clones with dual alpha chains were also detected. The major meta-clonotypes were represented in multiple mice.
Rhapsody analysis of Kb-reactive (PD-1hi), bystander (PD-1neg) or Kb self-tolerant T cells (from the Kb-transgenic H-2k strain 178.3) permitted profiling of a broader range of donor-reactive T cells. Comparison of V gene segment usage across the range of receptors expressed by PD-1neg cells from Sanger-sequenced or Rhapsody samples (502 or 1151 cells respectively) did not reveal any biases in the repertoire obtained based on the amplification method (Fig. 2a). TRAV14 and TRAV16 were over-represented among the PD-1hi cells compared to PD-1neg or Kb-tolerant cells while cells expressing TRAV9 and TRAV12 were less frequent (Fig 2b). The top 10 clones from each mouse accounted for 32% of all Kb-reactive (PD-1hi) cells. No clonal expansions were observed in the PD-1neg or Kb self-tolerant populations, with negligible overlap between TCR sequences from PD-1hi cells and these populations. Kb-reactive cells segregated into clusters corresponding to liver-resident or peripheral memory, precursor exhausted and proliferating cells, while PD-1neg cells displayed a central memory phenotype (Fig. 2c-d). Individual clonotypes showed distinctive gene expression patterns following activation.
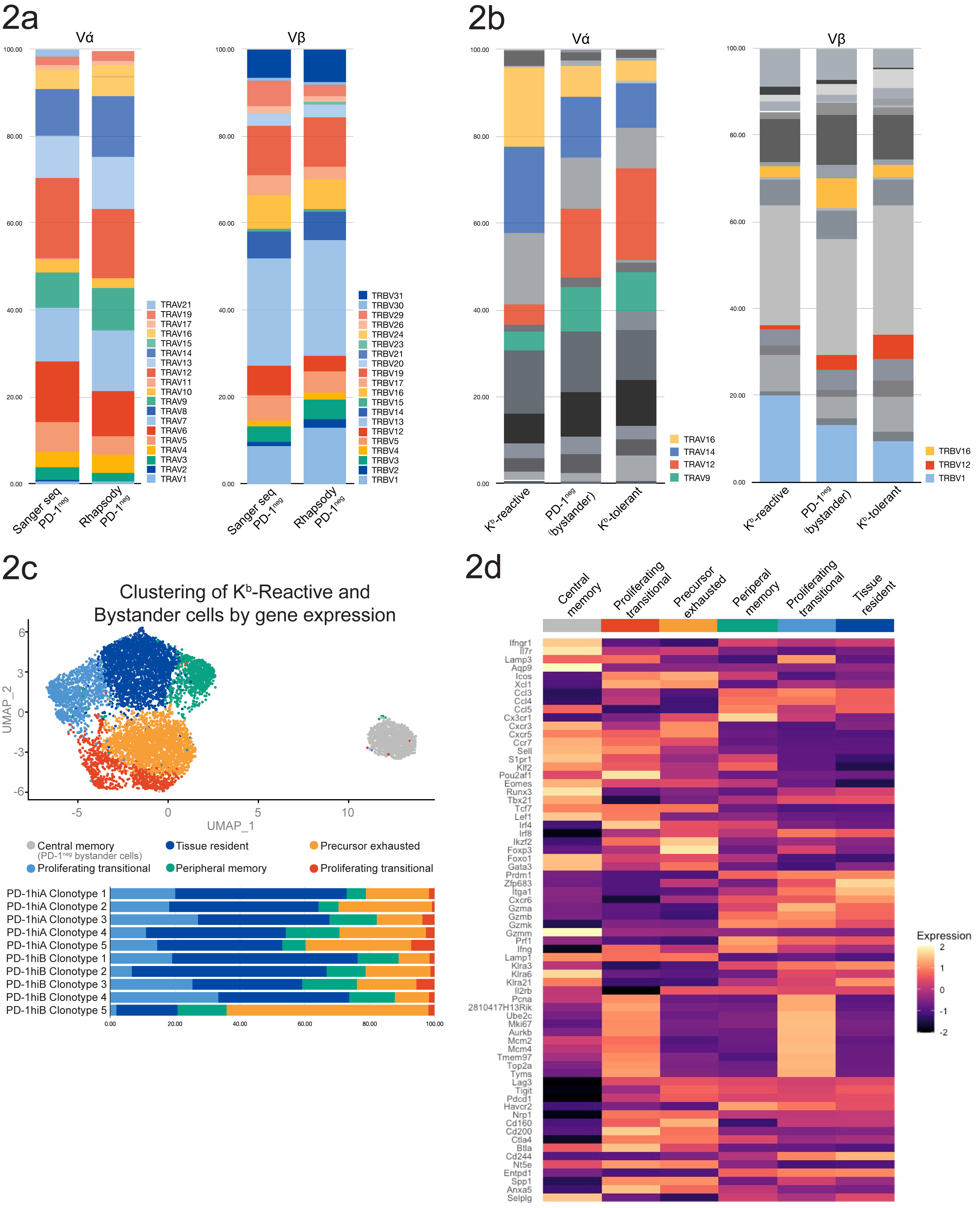
Conclusions: Directly-alloreactive CD8 T cell repertoires comprise families of closely-related clonotypes. Gene expression early during tolerance induction may determine the subsequent fate (deletion/exhaustion) of individual clones.
