Disease classification risk through machine learning algorithms – lessons learned from COVID-19
Xiao Zhang1, Maria Barros2, Maria Paula Gómez2, Entela Kondi2, Fritz Diekmann4, Chloë Ballesté2, Marián Irazábal2, P. Montagud-Marrahi4, E. Sánchez-Álvarez5, M. Blasco5, Martí Manyalich2, Ricard Gavaldà3, Pedro Ventura4, Jaume Baixeries1.
1Universitat Politècnica de Catalunya, Barcelona, Spain; 2International Cooperation, Donation and Transplantation Institute, Barcelona, Spain; 3Amalfi Analytics, Barcelona, Spain; 4Nephrology and Kidney Transplant Deparment, Hospital Clínic de Barcelona, Barcelona, Spain; 5Sociedad Española de Nefrología, on behalf of Spanish COVID Registry, Barcelona, Spain
Introduction: SARS-CoV2 virus disease registered more than 460,000,000 confirmed cases worldwide. Mortality due to SARS-CoV2 infection was higher in solid organ transplant recipients (SOT; 10-35% vs 5-7% in general population). We evaluated the utility of applying machine learning (ML) algorithms to a broad database (IDOTCOVID) aiming at developing tools which may be applied to other evolving clinical settings in the SOT field.
Method: We developed and compared 9 different ML models to predict the survival of SOT recipients infected with coronavirus. Models were run on IDOTCOVID, a worldwide database including 1400 patients from 78 transplant centers in 11 different countries between March 2020 and March 2021. Variables included both demographic and transplant related, as well as epidemiological, clinical manifestations, and treatment management of SOT’s with COVID-19. These prediction models include k-nearest neighbors (K-NN), linear regression, SVM with linear kernel, SVM with RBF kernel, two tree-based methods (decision tree, random forests), and three boosting methods (LogitBoost, AdaBoost, XGBoost). After a thorough evaluation, the best predictive machine learning model was used to predict patient survival status. A combined framework of machine learning algorithm and SHAP (SHapley Additive exPlanations) approach was built to provide the comprehensive interpretability of the model, including the discovery of important factors and an analysis of how individual important factors affect prediction outcomes and identification of the corresponding thresholds.
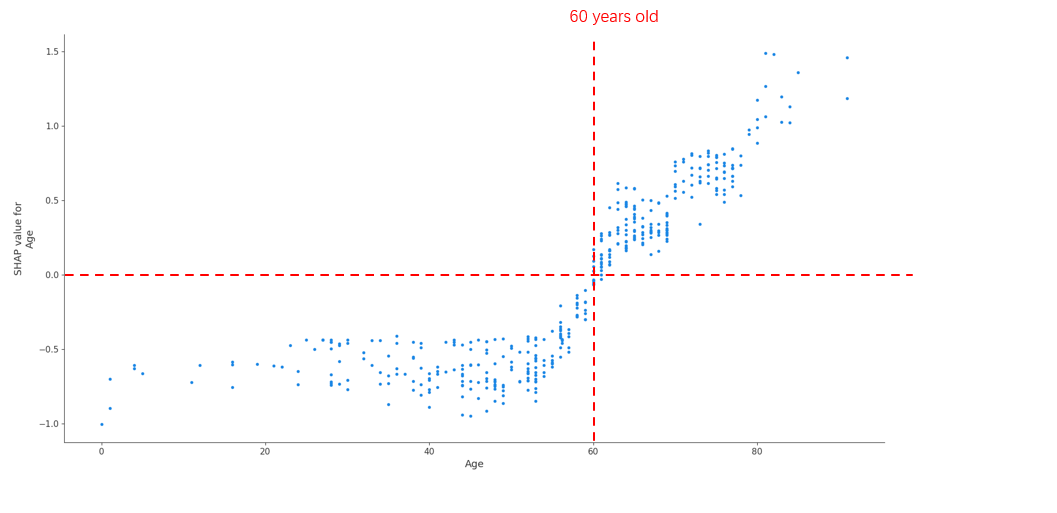
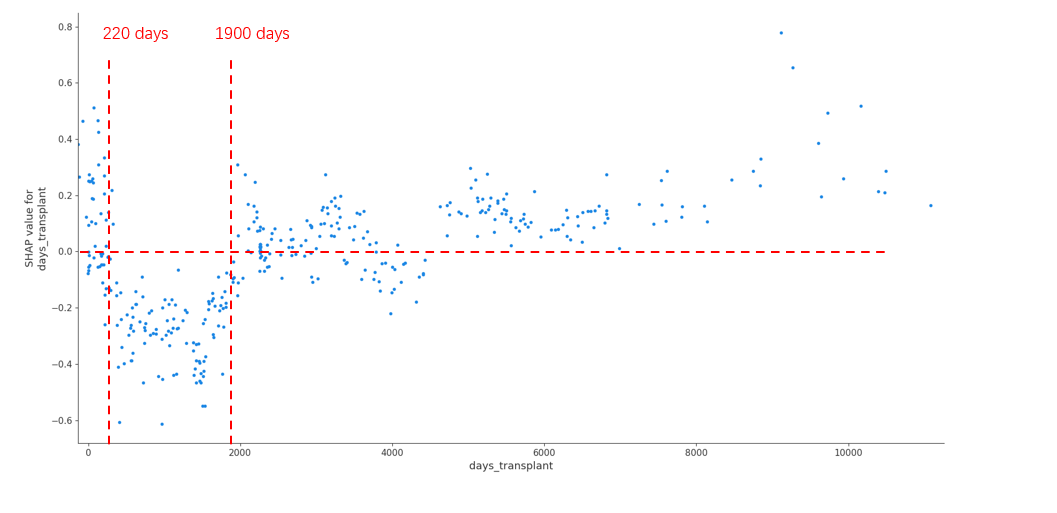
Result: Overall XGBoost achieved the best prediction performance for patient survival among all the algorithms, achieving an AUC of 0.842 (The Area Under the Curve). In the analysis of SHAP values for individual significant factors, three major categories emerged as significant for patient survival – infection period, recipient age, and transplant vintage. In comparing the survival of different patient subgroups, XGBoost performance was better for infections occurring during the first wave (until June 2020; AUC 0.842) than in subsequent waves (AUC 0.755). Conversely, XGBoost performance was better for younger patients (<60 years old; AUC 0.869) than for older patients (AUC 0.773) (Figure1). Finally, the SHAP analysis identified a U shape curve for patient survival according to time from transplantation, with recipients with an interval of less than 220 days or more than 5,2 years presenting an increased risk of death from COVID-19 disease (Figure 2).
Conclusion: The use of ML was able to accurately assess and predict the survival of recipients following SARS-CoV2 infection. XGBoost provided the best prediction results and may be regarded as an appropriate method for this task. The combination of prediction models and SHAP in broad up-to-date databases may help healthcare professionals identify and modulate important risk factors in evolving diseases.

right-click to download
