
Development and validation of an integrative dd-cfDNA system to predict allograft rejection: a population based study
Olivier Aubert1,2, Romain Brousse1, Cindy Ursule Dufait1, Juliette Gueguen1, Maud Racape1, Christophe Legendre1,2, Dany Anglicheau1,2, Carmen Lefaucheur1,3, Alexandre Loupy1,2.
1Paris Translational Center for Organ Transplantation, Paris, France; 2Kidney Transplant Department, Necker Hospital, Paris, France; 3Kidney Transplant Department, Saint Louis Hospital, Paris, France
Introduction: Post-transplantation patient care requires development and validation of non-invasive biomarkers to improve allograft monitoring and prevention from unnecessary biopsies. Preliminary reports have suggested the association of donor derived cell-free DNA (dd-cfDNA) with allograft rejection. However, there is no proof of its added value beyond standard of care patient management in large and deep phenotyped cohorts.
Method: 1210 concomitant evaluations of allograft histology, anti-HLA DSA and functional parameters between 2013 and 2018 were included corresponding to 637 evaluations in the derivation cohort and 573 in the validation cohort. dd-cfDNA was measured in plasma at the time of the evaluation. Diagnoses were assessed using Banff 2019 criteria. Parameters associated with kidney allograft rejection were assessed using uni- and multivariable logistic regression. We developed a risk model using the variables that were independently associated with rejection.
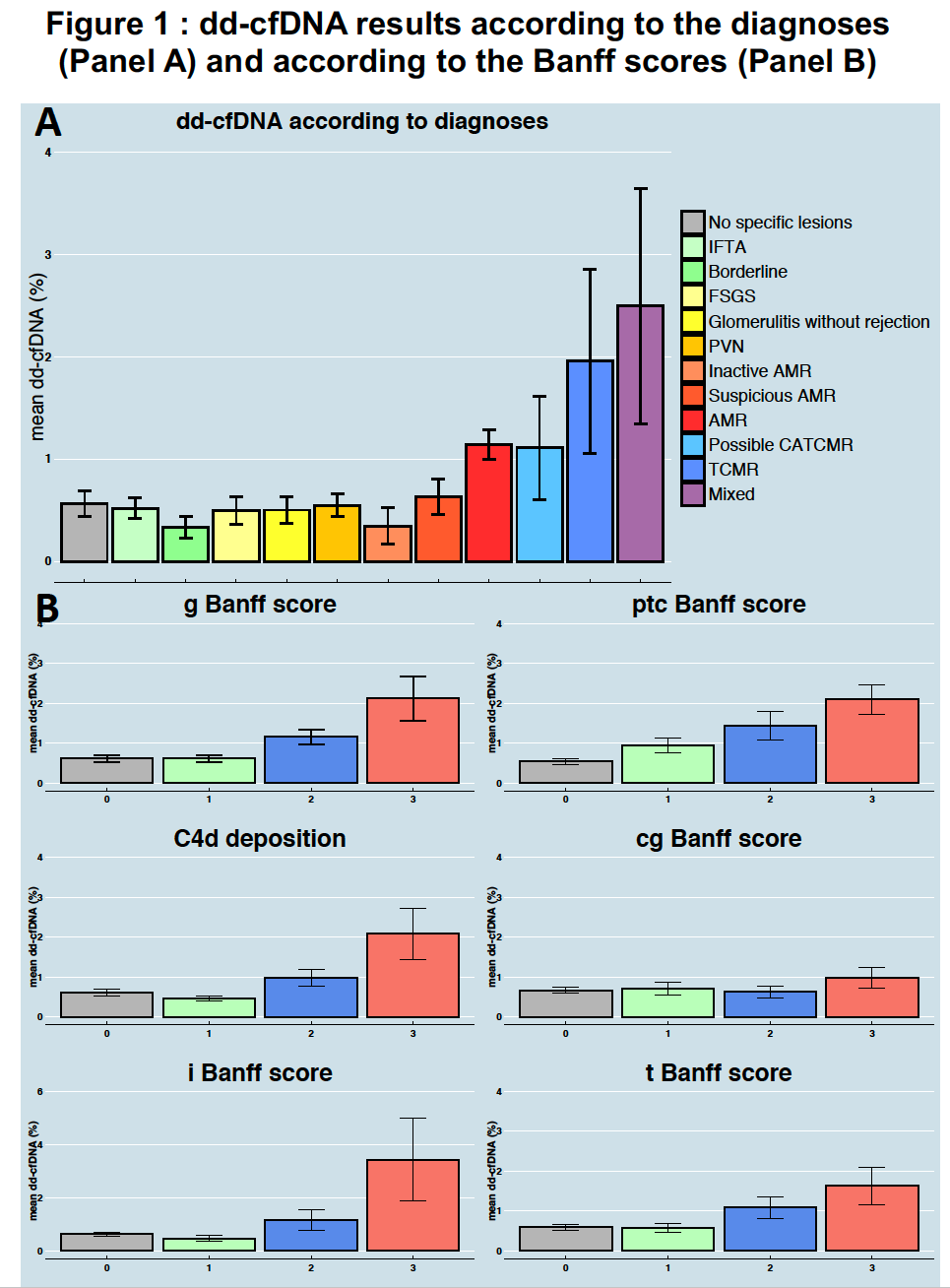
Results: Higher levels of dd-cfDNA were observed for AMR and TCMR or both compared to other diagnoses (Figure.1A). We found incremental dd-cfDNA levels with increasing Banff lesion scores for g, ptc, i, t, cg and C4d (Figure.1B) .There was no association of dd-cfDNA levels with allograft inactive lesions. In multivariable analysis, dd-cfDNA (p<0.0001) was associated with kidney allograft rejection independently of DSA (p<0.0001), eGFR (p=0.018), kidney allograft instability (p=0.013) and previous rejection (p<0.0001). Based on these parameters, we built an integrative idd-cfDNA model that showed good discrimination (AUC : 0.83), good calibration, and added value beyond a model without dd-cfDNA (AUC of the model without dd-cfDNA: 0.77 vs 0.83 for the integrative model; p<0.0001). We confirmed our results in the validation cohort with a good discrimination (AUC: 0.82) and a good calibration. This integrative score including the dd-cfDNA is being validated in Belgium and in the US.
Conclusion: We demonstrate the independent and added value of dd-cfDNA in addition to conventional features to predict rejection. This first integrative system shows improved performance for patient monitoring and could help physicians in decision-making process.

right-click to download
