Molecular characterization of progressive fibrosis in the liver by single-nuclei RNA sequencing
Thomas Rousselle1, Jennifer McDaniels1, Amol Shetty2, Elissa Bardhi1, Daniel Maluf3, Valeria Mas1.
1Department of Surgical Sciences, University of Maryland School of Medicine, Baltimore, MD, United States; 2Institute of Genomic Sciences, University of Maryland School of Medicine, Baltimore, MD, United States; 3Department of Surgery, University of Maryland School of Medicine, Baltimore, MD, United States
Introduction: Non-alcoholic fatty liver disease (NAFLD) is associated with an increase in inflammatory processes which potentially contribute to the exacerbation of fibrosis and development of cirrhosis. Hereby, we assess the use of single-nucleus RNA sequencing to identify the molecular mechanisms by which NAFLD progresses towards fibrosis at the single-cell resolution.
Methods: Single nuclei isolated from flash-frozen samples included normal, NAFLD, non-alcoholic steatohepatitis (NASH), and cirrhosis. Isolated nuclei were processed through a 10x Genomics Chromium Platform and analyzed via the Cell-Ranger pipeline. The Seurat package was used to generate clusters and cell identification. Cell types were analyzed for differentially expressed genes, which were further characterized by the predictive subcellular localization of the associated translated protein. Gene sets predicted to localize to the nucleus, cytosol, and mitochondria were individually explored for KEGG/GO-term enrichment and pathway analysis.
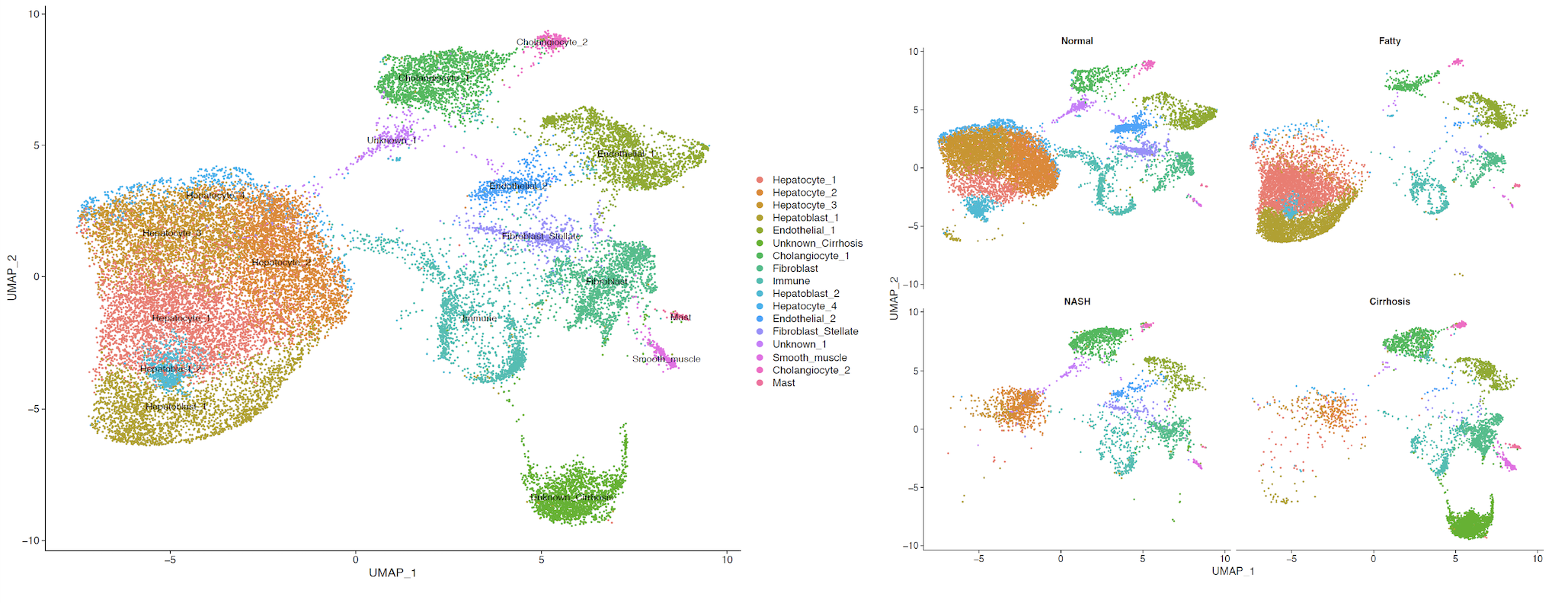
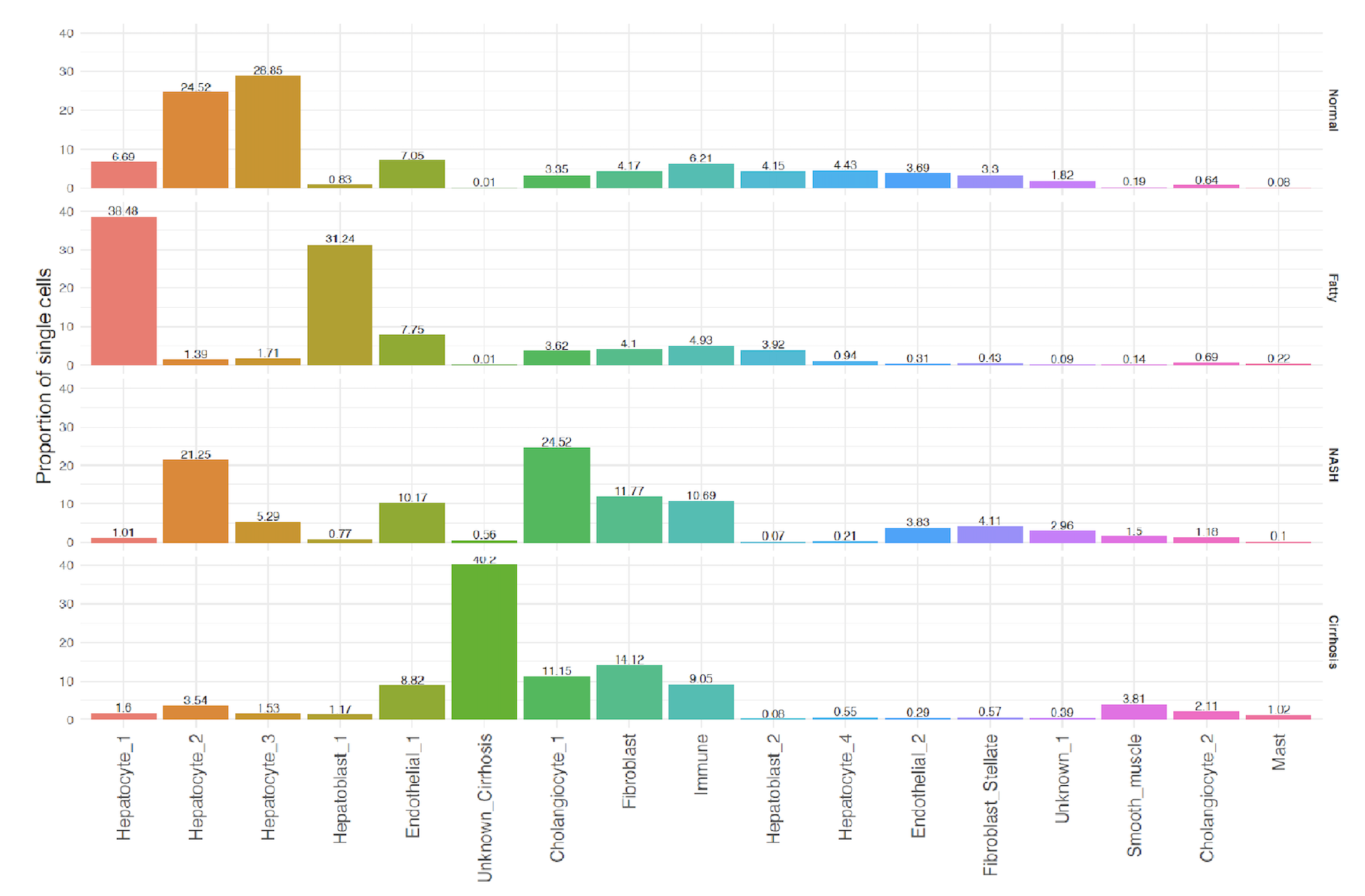
Results: Four hepatocyte (HEP) clusters and two hepatoblast (HpB) clusters were identified. HEP-1 and HpB-1 were found to be expressed almost exclusively in NAFLD. Gene clusters associated with mitochondria-localized gene products within the HpB-1 cell type were enriched for GO-terms specific to fatty acid oxidation and an upregulation of reducing power. In contrast, the HEP-1 cell type was found to have an increased reliance on amino acid metabolism, specifically the catabolism of branched chain amino acids by the upregulated activity of MCCC1, DBT, and HIBCH. In addition, gene clusters predicted to localize to the cytosol within the HEP-1 cell type were enriched for pathways specific for cellular response to oxidative stress. Taken together, these findings would suggest a progression of metabolic dysfunction from HpB-1 to HEP-1. In addition, these two cell types were also found to have a significantly higher concentration of genes associated with ECM production, including TIMP3, MMP15, ECM2, C1QTNF3, and SERPINE1.
Conclusion: In this study, we identify two cell types exclusively found in NAFLD which show an upregulated response to oxidative stress and fibrogenic mechanisms in light of progressive metabolic dysregulation. With continued progression, on-set of fibrosis lends to the eradication of hepatocytes and replacement of epithelial cells with fibrotic cells, thereby highlighting the cellular shift from NAFLD to Cirrhosis.

right-click to download
