Is there any role of combined use of gene expression profiling and donor derived cell free DNA in worsening renal allograft function
Ruchi Naik1, Walaa Dabbas1, Prerna Kumar1, Sajid Ansari1, Suman Setty2, Zahraa Hajjiri1, Ignatius Tang1.
1Transplant Nephrology , University of Illinois, Chicago, IL, United States; 2Pathology , University of Illinois, CHicago , IL, United States
Introduction: There are multiple noninvasive tests available to assess risk of renal allograft rejection. Donor-derived cell-free DNA (dd cf DNA) test is used to assess rejection in the settings of worsening renal allograft function while Gene profiling is used to assess the subclinical rejection in the settings of stable graft function. OmniGraf™ is a combination of Gene profiling and dd cf DNA tests and used for diagnosing possible subclinical rejection in the settings of stable renal allograft function. But not much has been observed in the settings of worsening renal allograft function. Here we are presenting our study aiming to correlate OmniGraf™ result with the biopsy outcomes.
Methods: We selected 40 patients who had a biopsy either for cause or for protocol along with OmniGraf™ and serum creatinine checked around the same time of the biopsy. Patients were divided into 2 major groups, 1.protocol, and 2. For-cause biopsy, later subdivided into 4 groups depending on the OmniGraf™ result combinations. TX is considered No Rejection, Non-TX is considered increased risk of subclinical rejection.TRAC is the dd cf DNA test, High TRAC more than 0.7% is considered increased risk of rejection.
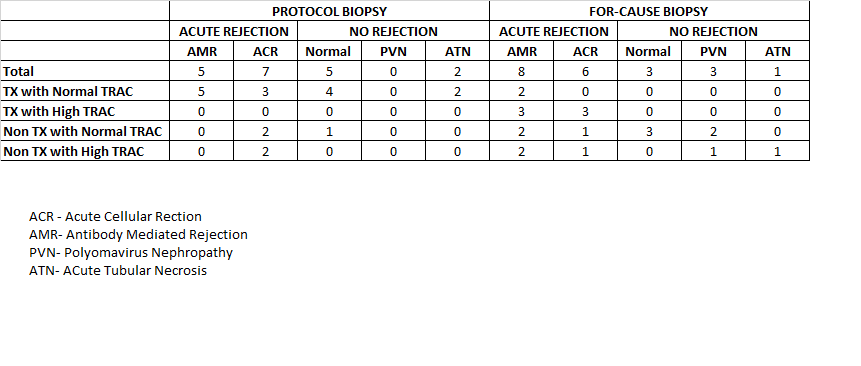
Result: In the protocol biopsy group 12 out of 19 patients who had TX with normal TRAC had rejction on the biopsy.The normal result of OmniGraf™ TX with normal TRAC was supposed to predict no rejection, so if biopsy was not performed it might have missed 57% of rejection.This suggest that the normal result does not predict absence of rejection. The subgroup NonTX with Normal TRAC had 66% patients showing rejection,showed 100% rejection. This suggests that Non TX, irrespective of TRAC level is good in predicting possible rejection, irrespective of the type of rejection.In the for-cause biopsy group, the subgroup 2 of 2 patients with TX and normal TRAC had rejection on biopsy. Normal result of OmniGraf™ is not reliable to rule out rejection. The subgroup TX with high TRAC is good in capturing rejection for 6 out of 6 patients. This suggest that Tx may not be useful in this group in predicting rejection as dd cf DNA test, alone might have been able to predict the rejection. The subgroup Non TX with normal TRAC is showing 37.5 % patients, positive for rejection on biopsy while NON TX with high TRAC group has 60% of patients with rejection. This suggest that Non-TX part is good in predicting increasing risk of rejection in both groups irrespective of the High TRAC.
Conclusion: Checking a single OmniGraf™ test to evaluate the risk without doing biopsy will miss subclinical rejection.It is probably more useful in serial monitoring of OmniGraf™ than one single value to capture subclinical rejection. Our data are limited to suggest clinical or statistical relevance, It would be useful to evaluate these results on a larger scale to understand the usefulness of this test in kidney transplant recipients with worsening renal allograft function.

right-click to download
