Comparison of HLA compatibility algorithms to predict long-term survival and CLAD following lung transplantation
Steven Hiho1,2, Bronwyn Levvey2, Greg Snell2, Glen Westall2, Lucy Sullivan2,3.
1Lung Transplant Service, Department of Respiratory Medicine , Alfred Hospital, Melbourne, Australia; 2Victorian Transplantation and Immunogenetics Service, Australian Red Cross LifeBlood, Melbourne, Australia; 3South Australian Transplantation and Immunogenetics Service, Australian Red Cross LifeBlood, Adelaide, Australia
Introduction: Outcomes following lung transplantation (LTx) remain poor, despite recent advances in sequencing technology and the development of multiple algorithms defining immunological compatibility between recipient and donor. No consensus has been achieved regarding the best approach to define HLA compatibility in LTx. Here, we compared numerous available HLA compatibility approaches, including T- and B-cell epitopes, electrostatic charge and amino acid mismatching, using a high resolution typed, well characterised cohort, to determine which approach best predicts outcomes following LTx.
Methods: In this retrospective single-center study, 277 donor-recipient transplant pairs were retrospectively HLA typed by Next generation sequencing (NGS) for all HLA loci. (HLA-A-DPA). HLA mismatching was defined using HLAMatchmaker eplets (epMM), HLA-EMMA epitopes (EMMA), PIRCHE T-cell epitopes (PIRCHE), electrostatic differences (EMS) and amino Acid mismatches (AAMM). Associations with mismatching scores generated by the different algorithms with CLAD (BOS/RAS) and overall survival were calculated using adjusted cox proportional modelling and Kaplan-Meier survival curves to demonstrate time to outcomes (SPSS v.21).
Results: Lower HLA-class II compatibility was associated with increased overall survival for all algorithms, with lower HLA-DR, -DQ and –DQA mismatches generated by PIRCHE and epMM independently associated with increased survival. Lower HLA-DR compatibility was significantly associated with increased time to RAS for all algorithms. Lower epMM and PIRCHE HLA-DQA was also associated with increased time to RAS, however the algorithms that did not separate DQA and DQB, did not reach significance.
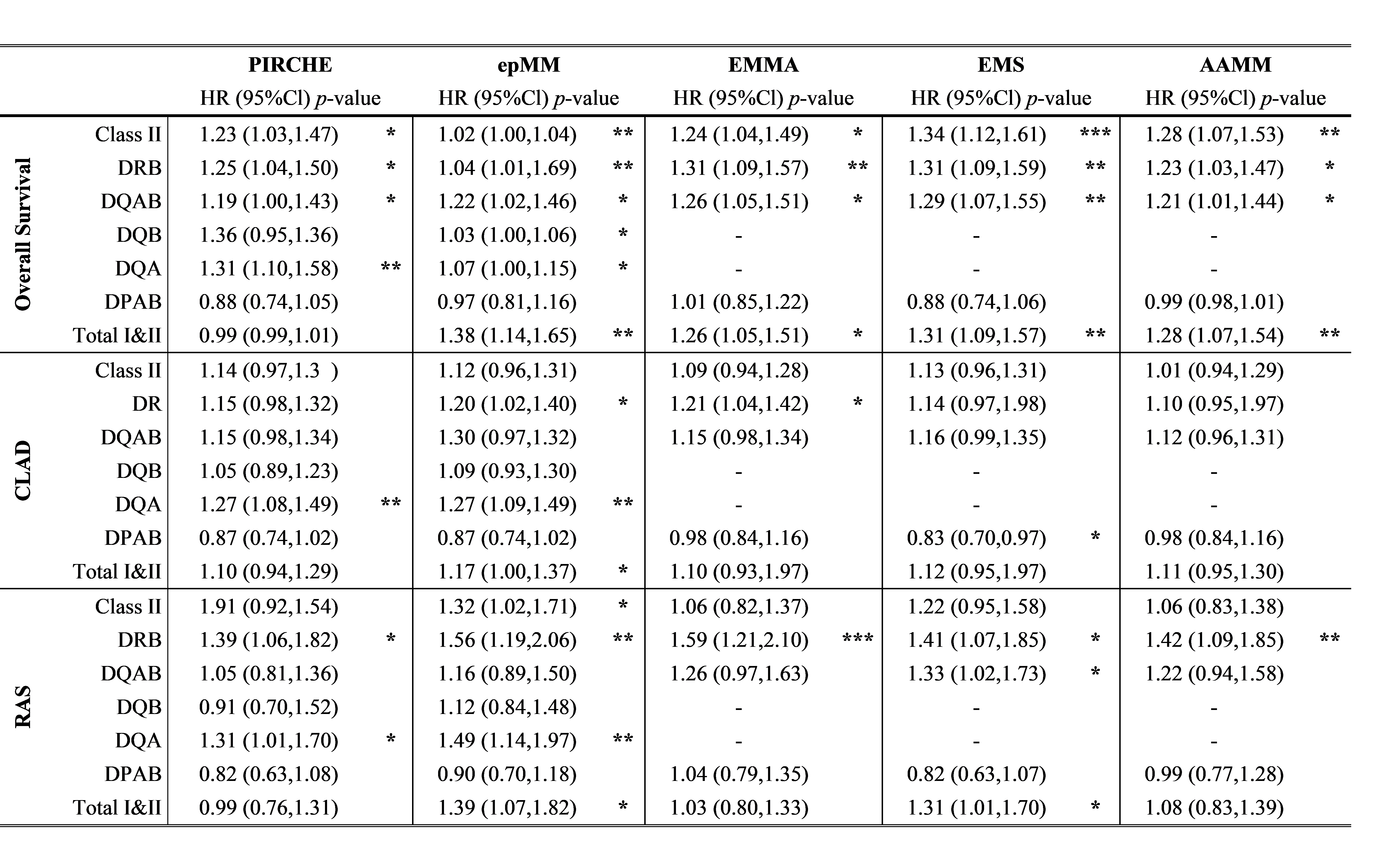
Conclusion: Lower class II mismatching, specifically for HLA-DR and DQ, for all approaches associated with improved graft and patient survival. The PIRCHE and HLAMatchmatcher algorithms that separate compatibility based on individual alleles/chains provide a higher degree of granularity, which may be useful to select the best matched recipient. Regardless, reducing the level of HLA mismatching, in either T- or B-cell epitopes, electrostatic differences or amino acid would improve outcomes following LTx and potentially guide immunosuppression strategies.

right-click to download
