Chimerism and bidirectional alloresponses in patients receiving lung transplantation
Jianing Fu1,2, Wenyu Jiao1, Kortney Rogers1, Constanza Bay Muntnich1, Arnold Valena3, Katherine D. Long1, Joseph Costa4, Steven Russum4, Zhou Fang1, Nichole Danzl1, Luke Benvenuto2, Joshua Sonett4, Philippe Lemaitre4, Frank D’Ovidio4, Selim Arcasoy2, Megan Sykes1,2,4,5.
1Columbia Center for Translational Immunology, Columbia University, New York, NY, United States; 2Department of Medicine, Columbia University, New York, NY, United States; 3NewYork-Presbyterian Hospital, Columbia University, New York, NY, United States; 4Department of Surgery, Columbia University, New York, NY, United States; 5Department of Microbiology & Immunology, Columbia University, New York, NY, United States
Introduction: Long-term outcomes after lung transplantation (LuTx) are suboptimal, with 5-year survival < 60%. Graft rejection is a major complication limiting success. Lung grafts carry large numbers of T, B and antigen-presenting cells, leading to two-way alloresponses: graft-vs-host (GvH) and host-vs-graft (HvG). Despite the importance of T cells in driving alloresponses, their dynamic repopulation, clonal distribution and alloreactivity after LuTx are largely unknown.
Methods: Serial bronchoalveolar lavage (BAL) provides a unique opportunity for temporal analysis of immune cells in lung grafts. We utilized a combination of flow cytometry that includes HLA-specific antibodies to distinguish donor- and recipient-derived cells, with high-throughput sequencing of donor and recipient T cells within pre-Tx lymphoid tissues and identification of alloreactive T cell clones in the GvH and HvG directions in mixed lymphocyte reactions (MLRs). Data generated from patients (Pts) 1 and 4 were reported here, where we had a relatively complete tissue collection, including the donor and recipient tissues pre-Tx to perform MLRs and 2-4 time points of BAL and lung biopsy specimens post-Tx to track chimerism and clonal distribution.
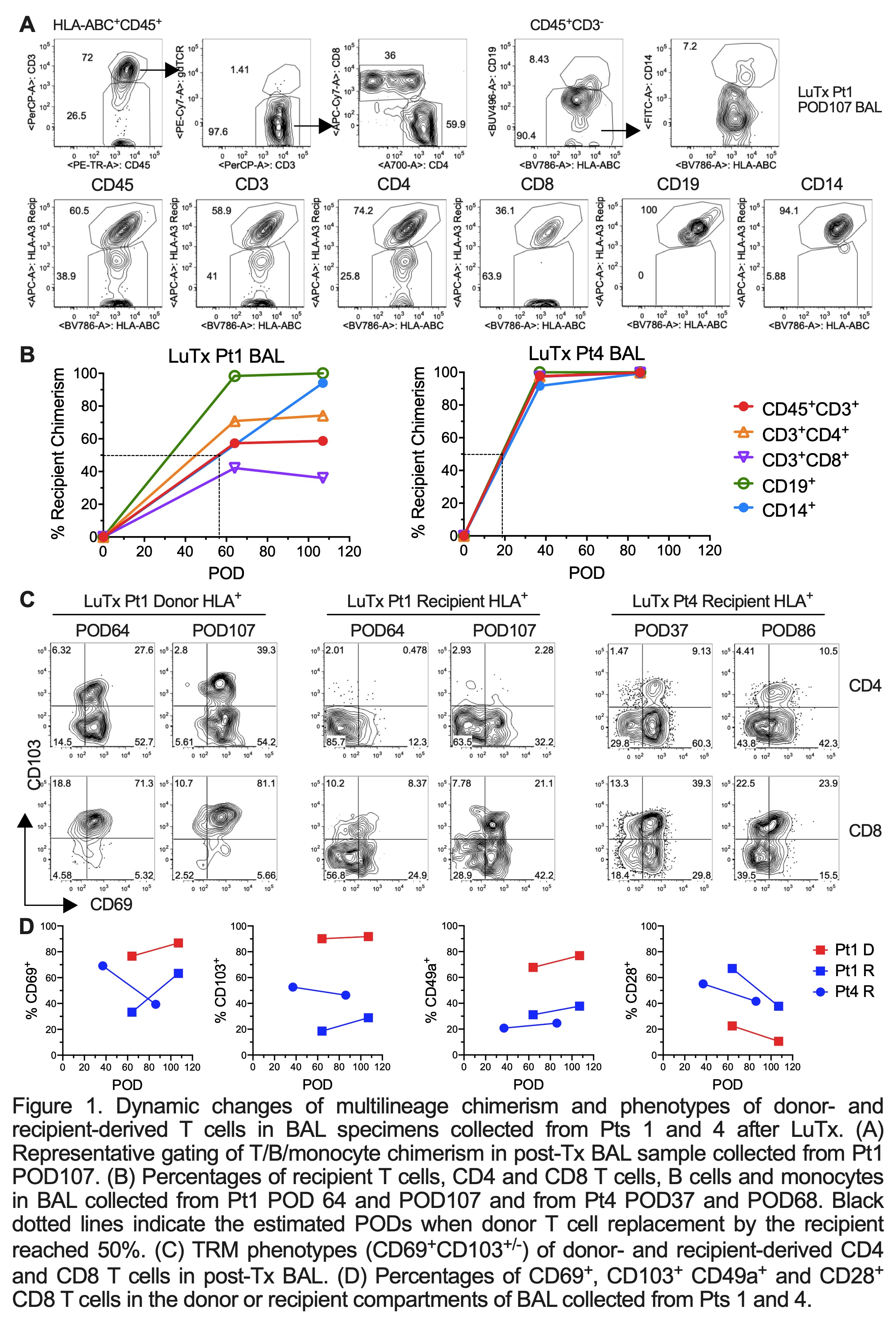
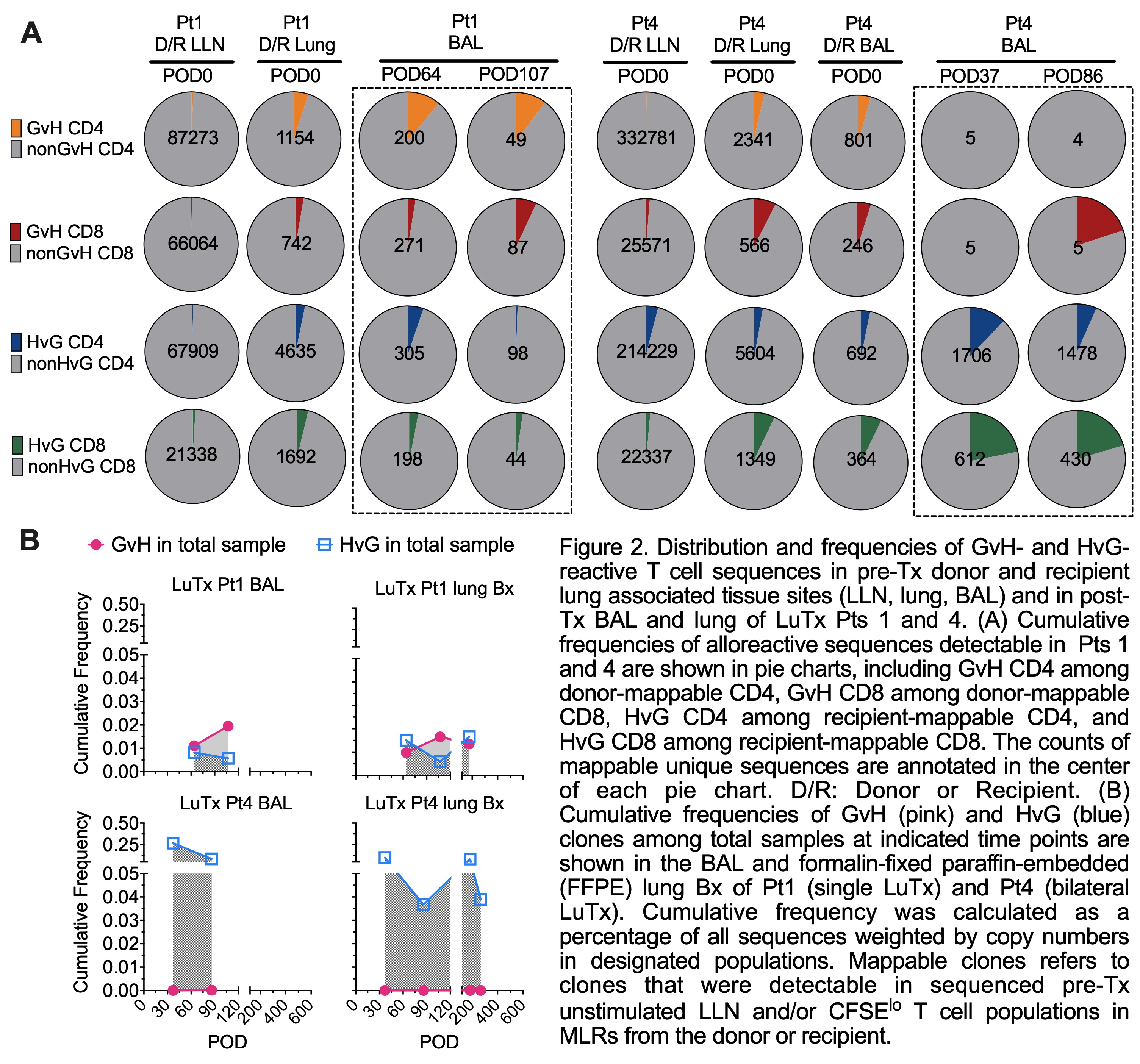
Results: Donor T/B/monocytes in BAL post-Tx were gradually replaced by the recipient (Fig.1A-B). The turnover dynamics were faster in Pt4 compared to Pt1, especially for T cells. Donor CD8 T cells showed persistent high expression of TRM markers (CD69, CD103 and CD49a) and low expression of the effector T cell (Teff) marker CD28 (Fig.1C-D). However, recipient graft-infiltrating CD8 T cells gradually downregulated CD28 during the process of acquiring TRM features, indicating a phenotypic transition from circulating Teffs to lung-resident TRMs. An enrichment of HvG- compared to GvH-reactive sequences was observed in Pt4 among both pre-Tx mappable repertoires (Fig.2A) and all sequences in BAL post-Tx (Fig.2B). However, an opposite trend was shown in Pt1 (Fig.2). The patterns of GvH and HvG clonal frequency in lung biopsies were quite similar as observed in BAL in both patients (Fig.2B). Despite obvious differences in the turnover dynamics and alloreactive repertoire of T cells in BAL and lung of these two patients, histology on the lung transbronchial biopsies taken on the same day reported no signs of rejection for either.
Conclusions: Our data suggest that cellular and clonal repertoire changes may have already happened in lung allografts when histology is not diagnostic of rejection. The association among the dynamic replacement of T cells in lung allografts, differences of donor graft lymphoid cell load, and the presence and dominance of GvH and HvG-reactive T cell sequences in association with graft outcomes will be further investigated in an expanded cohort, revealing underlying mechanisms of graft acceptance and rejection and eventually help predict graft outcomes and develop novel strategies to promote tolerance after LuTx.

right-click to download
